rxncon - the reaction-contingency language
rxncon ("reaction-con") is a framework to build, visualise and model cellular signal transduction networks.
The rxncon language makes it easy to formalise detailed mechanistic knowledge on cellular signal transduction. The language is computer readable, and the rxncon compiler can convert the knowlede base into a range of graphical and executable outputs.
We are inspired by the comprehensive wall charts - and later genome-scale models - that revolutionised our understanding of cellular metabolism. With rxncon, we strive to make the same possible for cellular signal transduction.
We finalised a major overhaul of the language, the interpretation engine and the export features and are happy to present rxncon 2.0, the second generation rxncon language with an increased expressiveness and precision.
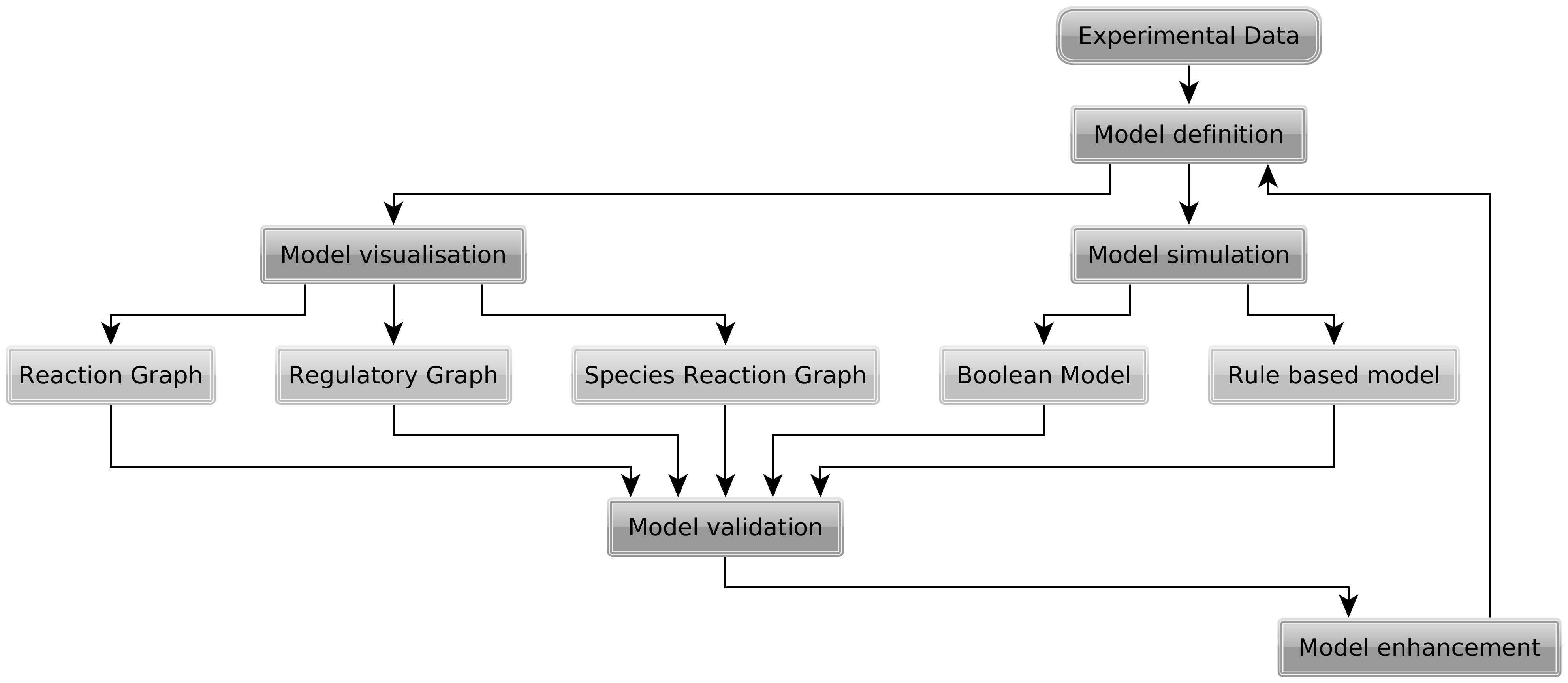
A model formulated in the rxncon language is a knowlede base. The knowledge can be visualised in a range of graphical formats, or compiled into executable models. We have developed a parameter-free simulation logic that can be used for network validation and qualitative simualtion. Alternatively, the network can be converted into a rule-based model that requires parametrisation before simulation. The higher level "biological" language and multiple compilation targets make it easier to develop models. Automatic visualisation - with layout reuse - and model generation greatly facilitate an iterative model development - visualisation - validation process.